Data analysis
FragMAXapp offers a variety of pipelines for data analysis. The options are presented in the Data analysis tab.
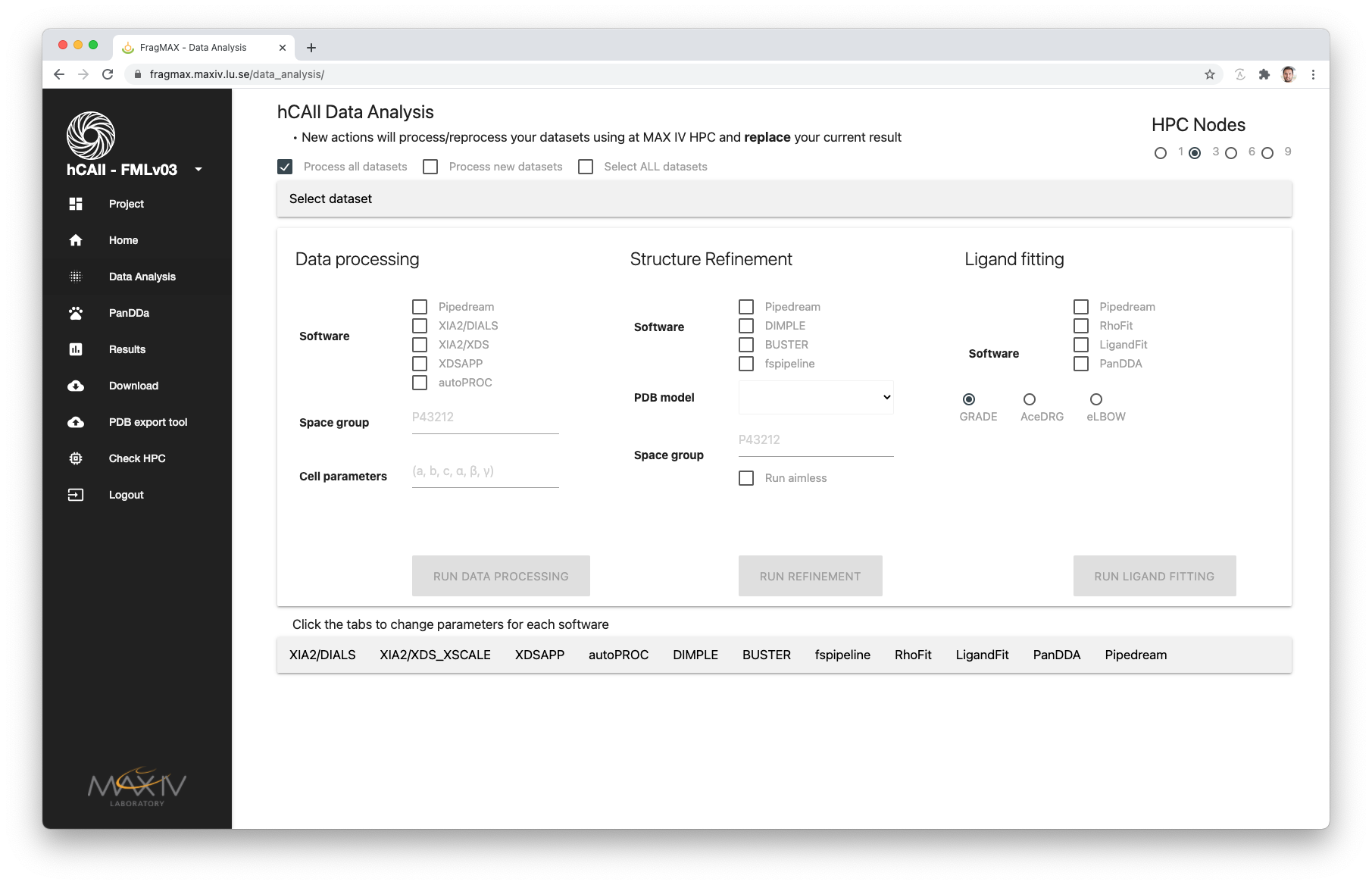
On the top left corner, checkboxes gives a quick selection of datasets as follow:
| Option | Selection |
|---|---|
| Process all datasets | All datasets available in the provided shifts |
| Process new datasets | Selects all datasets that is marked in yellow in the home page (A.K.A. datasets that was not analysed previously |
| Select ALL datasets | This option will mark all checkboxes in the Selected datasets tab |
| No checkbox checked | The datasets selected in the Selected datasets will be used |
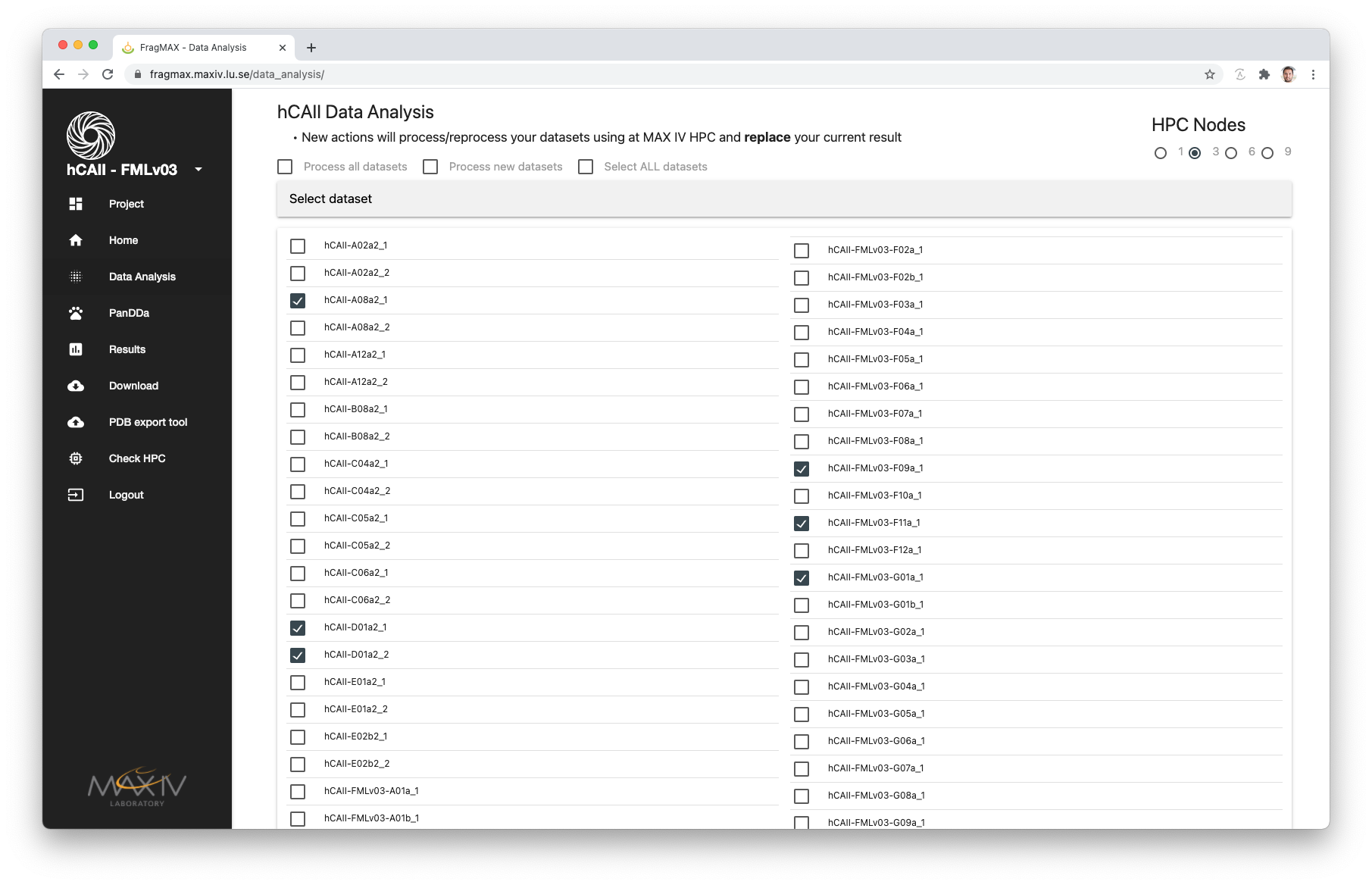 Dataset selection tab
Dataset selection tab
Once the minimum information is provided to a give step, the referred button will be enabled.
| Step | Option | Expected input | Effect | Remarks | Required |
|---|---|---|---|---|---|
| Data processing | Software | Select the checkboxes | Uses the method on all selected datasets | Every new analysis will replace previous results | Yes |
| Space group | e.g.: P43 21 2 | The SPG provided will be used during data processing | XDSAPP requires Space group number instead, e.g.: 96 | No | |
| Cell parameters | e.g.: (66,66,100,90,90,90) | The Cell parameters will be used during data proessing | Please check if the selected software supports only SPG or Cell Param. Example: XDSAPP will required both, XIA2 can take only one | No | |
| Structure refinement | Software | Select the checkboxes | Uses the method on all selected datasets | Every new analysis will replace previous results | Yes |
| PDB model | Select one of the project’s PDB models | Selected model will be used for structure refinement | PDB models should be added through Project definitions | Yes | |
| Space group | e.g.: P43 21 2 | The SPG provided will be used during structure refinement | Yes | ||
| Run aimless | Select the checkboxes | Run aimless prior to structure refinement | Fixes some issues regarding alternative SPG during data processing | No | |
| Ligand fitting | Software | Select the checkboxes | Uses the method on all selected datasets | Every new analysis will replace previous results | Yes |
| SMILES to CIF | Select the option | Convert fragment SMILES to CIF using selected method | Fragments can be updated through Library view | Yes |
This options can be parsed in the screen below
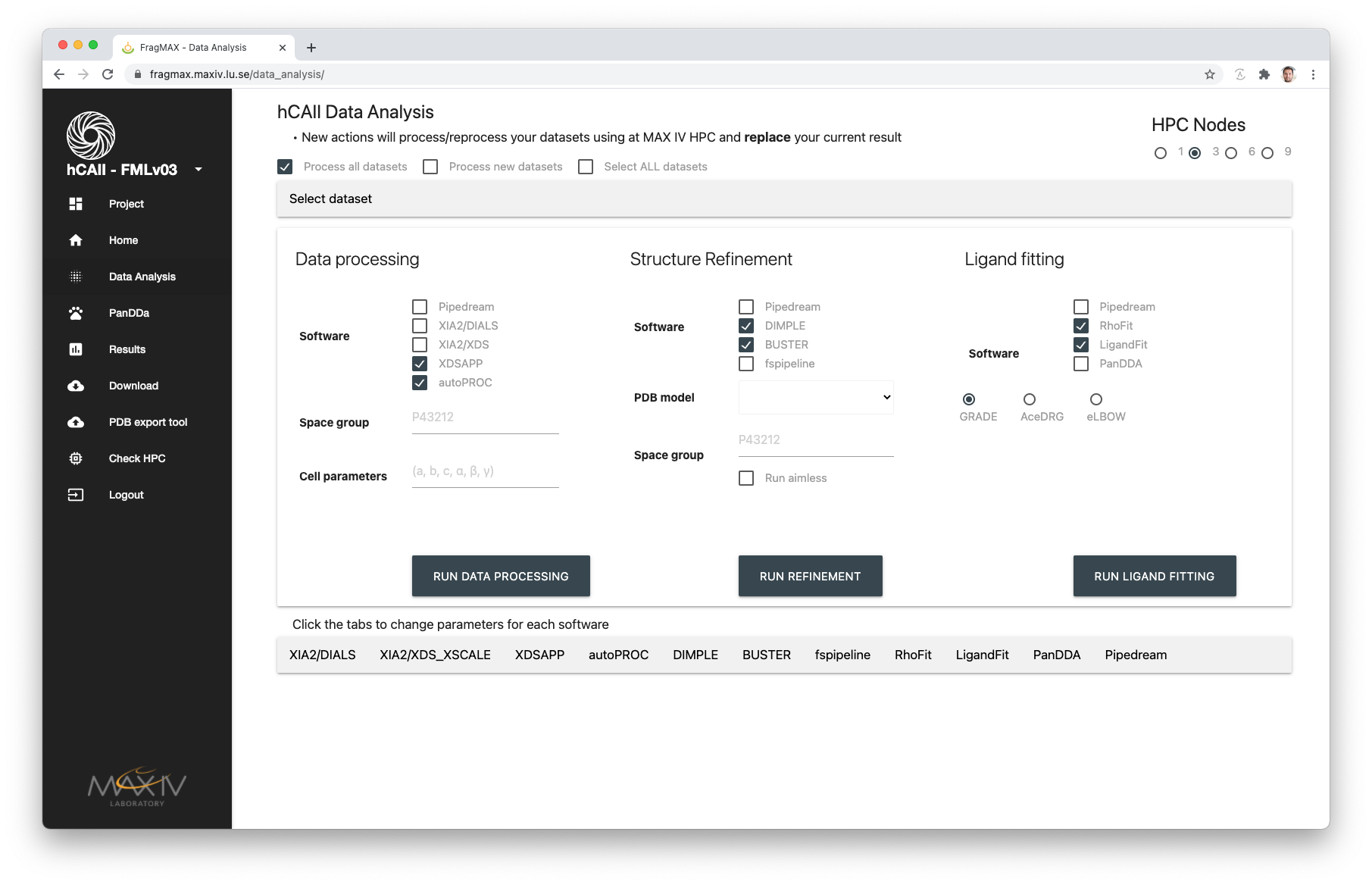
Custom parameters
For each method, custom paramters are available.
The tabs on the bottom of the page will provide some extra pre-selected customisations and one extra field, Custom parameters.
In the Custom Parameters, it is possible to pass anything supported by the method.
Example:
DIMPLE
* -M0
* will be used to do Molecular replacement always
* -M0 --slow
* will do MR and extra cycles of refinement
NOTE: The custom paramteres should be passed as they would in the command line version of the method. Links for the method documentation is provided below the input box.
Pipedream
Selecting GlobalPhasing Pipedream will open the referred tab for extra inputs.
Please not that information passed to Pipedream will ONLY come from the tab. Dataset selection works as usual, from the top checkboxes selection.