Login Credentials
Username and password are provided by the hosting site.
At MAX IV Laboratory, this is provided by the User Office and it is the same across the facility

Project Setup
If you don’t have any projects yet, this will be your landing page
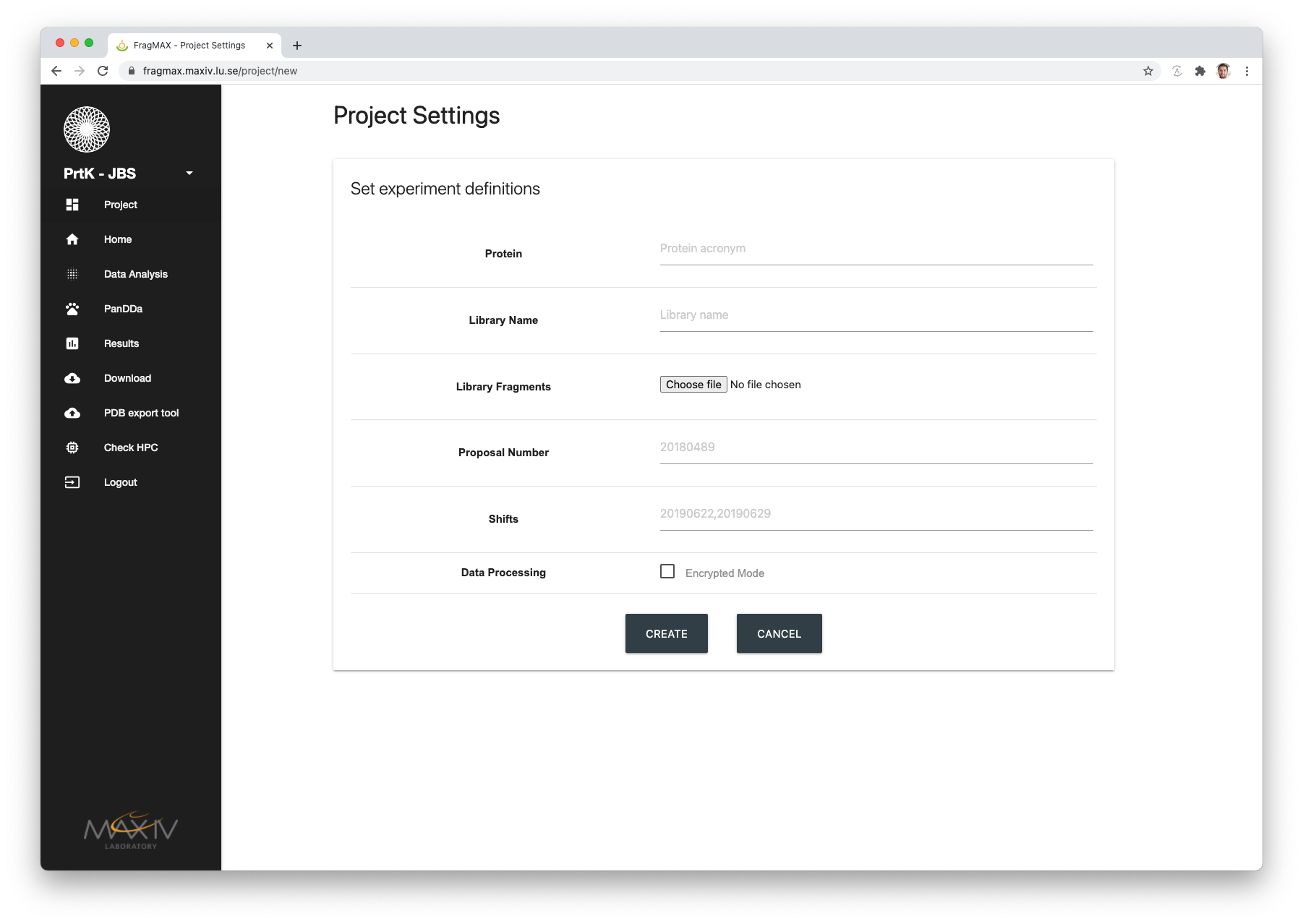
| Field | Expected input |
|---|---|
| Protein | Protein name as declared in ISPyB/EXI. Example |
| Library Name | Fragment library name as declared in ISPyB/EXI |
| Library fragments | CSV file containing fragments definitions. See how to create this file HERE |
| Proposal number | FragMAX project proposal number |
| Shifts | If the site you are using FragMAXapp supports it, input the visits according to local format (e.g.: MAX IV -> 20201022) |
| Data Processing | Enable/Disable encryption mode for data processing |
After providing necessary information, the project will enter the Setting up mode. This mode will lock the use of the project until:
- Data collection files are read and parsed to FragMAXapp database
- CIF files are generated from input SMILES
- Data from beamline autoprocessing is imported to FragMAXapp
Update or remove projects
Click in the top left-hand corner of the page and then Manage projects in the upcoming menu.
Clicking in one of the available projects, it is possible to ammend definitions of a project or delete it. Deleting projects will not instantly remove the data from the disk.
Library definitions
- Your library file must be in CSV format.
- Each line of your file describes one fragment as
- fragmentID,SMILES
Example:
mylibrary.csv
A01a,O=C1N[C@@H](CO1)C1=CC=CC=C1
A01b,CN1CCCCS1(=O)=O
A02a,CC1=CC=C(S1)C1=CC(=NN1)C(O)=O
A02b,CC1=CC(C)=NC=N1
Standard fragment libraries definitions
FragMAXlib
FragMAXlib is formulated via in-house fragment collection of FragMAX. FragMAXlib is an XFS-focused library, specifically designed for FragMAX operations. The library is in a constant refinement cycle and is periodically updated, depending on the fragment performance in screening experiments, new chemotypes in a commercial chemical space and platform updates. FragMAXlib compounds are selected on the basis of their fragment-likeness and synthetic tractability. Currently, the library has 110 entries.
| Descriptor | min | max | mean |
|---|---|---|---|
| Mw | 108 | 269 | 158 |
| Num heavy atoms | 8 | 18 | 11.2 |
| SlogP | -0.7 | 2.6 | 1 |
| Num. H-bonds acc. | 1 | 5 | 2.5 |
| Num. H-bonds don. | 0 | 2 | 0.9 |
| Num. rot. bonds | 0 | 3 | 1 |
Frag Xtal Screen
Frag Xtal Screen is a commercially available fragment library. Fragments are formulated in a solid form, allowing to perform screening experiment even if a crystal system does not tolerate organic co-solvents, e.g. DMSO. The library consists of 96 fragment-alike compounds and crystallization additives.
Huschmann et al. 2016. “Structures of endothiapepsin-fragment complexes from crystallographic fragment screening using a novel, diverse and affordable 96-compound fragment library”
F2X-Entry
From the F2X-Universal library we made a sub-selection creating a 96 fragment screen that is still maximally diverse and that we provide in ready-to-use plates to the users. So screens can be carried out on-site or in the user’s home lab. This screen has currently been validated with several test cases and showed high hit rates (~20% on average, 6 campaigns). Further campaigns are ongoing.
Wollenhaupt & Metz et al. 2020. F2X-Universal and F2X-Entry: Structurally Diverse Compound Libraries for Crystallographic Fragment Screening
FragMAX naming template
FragMAXapp will look for data in the user’s directory matching the following criteria
- ProteinName-LibraryName-FragmentID_master.h5
- ProteinName-LibraryName-FragmentID_?????.cbf
Name restrictions
| Field | Restriction |
|---|---|
| ProteinName | - _ , / " " * [](){} |
| LibraryName | - _ , / " " * [](){} |
| FragmentID | - _ , / " " * [](){} |
| Overall | The final combination cannot be the same |
Examples:
| Final sample name | |
|---|---|
| Right | PrtK-XtalFragScreen-A01 |
| PrtK-XtalFragScreen-0001 | |
| Wrong | PrtK_XtalFragScreen.A01 |
| PrtK XtalFragScreen-A01 | |
| PrtKXtalFragScreenA01 |
PDB upload
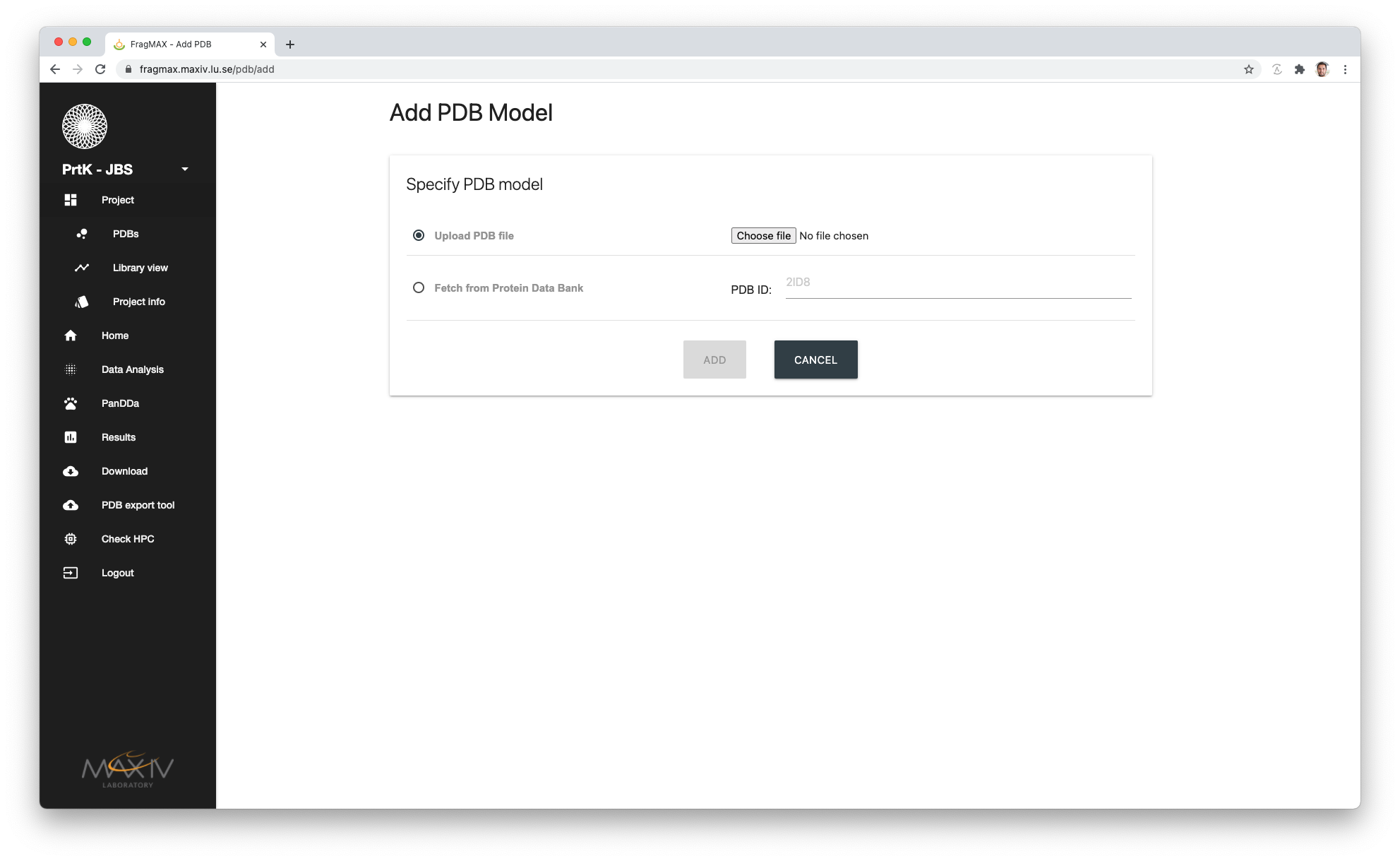
Under the menu
- Project
- PDBs
It is possible to add multiple PDBs to your project from your local computer.
Alternatively PDB can be directly fetched from RCSB website by providing the PDB ID
For each file uploaded, three models will be generated:
- file.pdb
- The exact copy of provided PDB
- file_noHET.pdb
- Removing HETATM tags from the PDB. The HEADER and REMARK is kept.
- file_txc.pdb
- If your uploaded file contains 2 or more chains, a trimmed crystallographic components version of your file is generated using phenix.sculpt_ensemble
The models can be downloaded and/or replaced at any time during the project.
Library overview
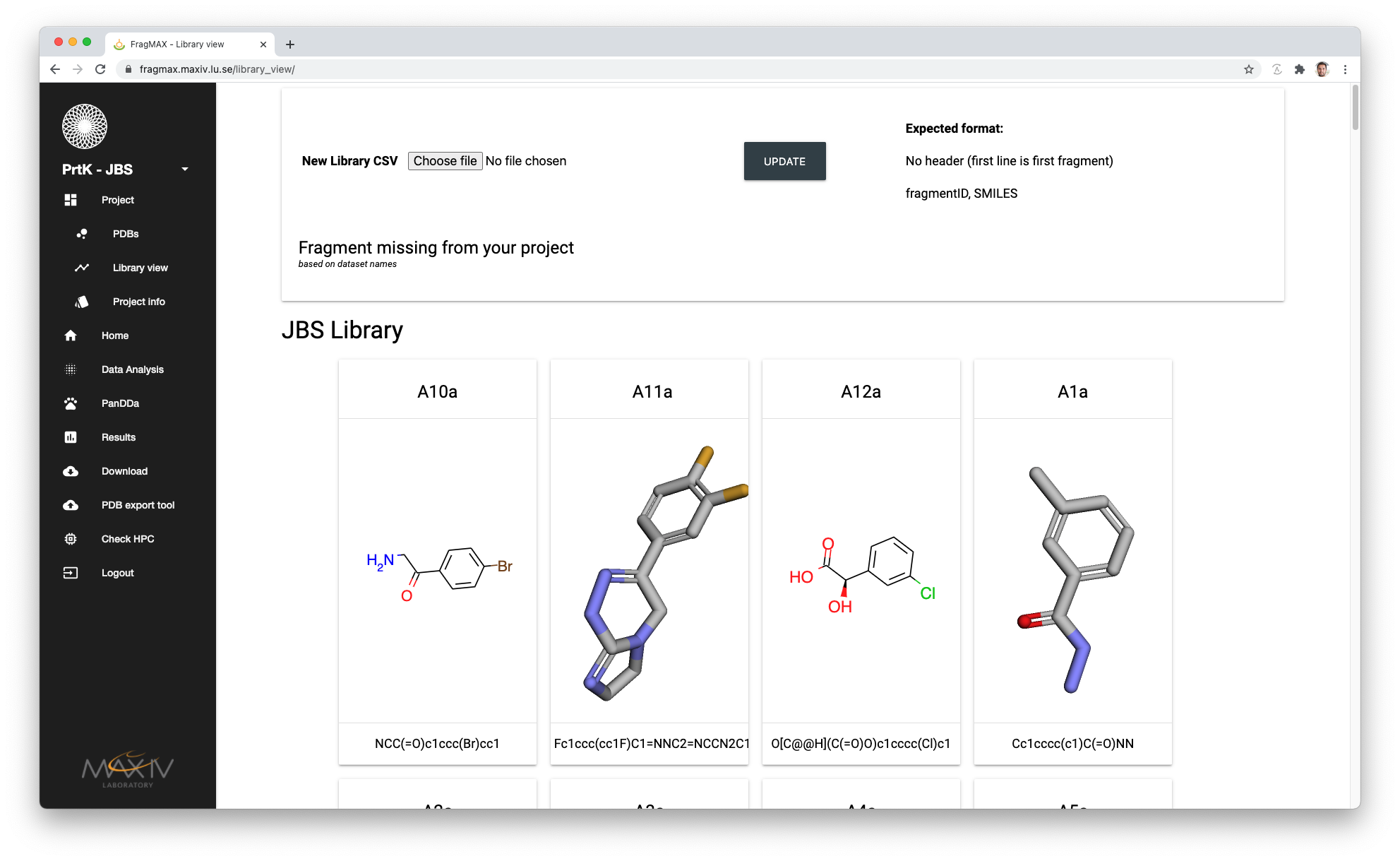
Under the menu
- Project
- Library view
FragMAXapp will display the current loaded library. Missing fragments (based on sample names and fragIDs provided) will be displayed in red until uploaded to the project.
Uploading new library can be done in two ways:
- CSV file with all definitions.
- This will replace all current SMILES in FragMAXapp database with the information from the CSV file.
- CSV file with one or more fragments.
- This will update or add only the fragments in the file, leaving all other unchanged.